The goal of xplainfi is to collect common feature importance methods under a unified and extensible interface.
It is built around mlr3 as available abstractions for learners, tasks, measures, etc. greatly simplify the implementation of importance measures.
Installation
Install xplainfi from CRAN:
install.packages("xplainfi")Or install xplainfi from R-universe:
install.packages("xplainfi", repos = c("https://mlr-org.r-universe.dev", "https://cloud.r-project.org"))The latest development version of xplainfi can be installed with pak:
# install.packages(pak)
pak::pak("mlr-org/xplainfi")Example: PFI
Here is a basic example on how to calculate PFI for an untrained learner and task, using cross-validation for resampling and computing PFI within each resampling iteration 10 times on the friedman1 task (see ?mlbench::mlbench.friedman1).
The friedman1 task has the following structure:
\[y = 10 \sin(\pi x_1 x_2) + 20(x_3 - 0.5)^2 + 10x_4 + 5x_5 + \varepsilon\]
Where \(x_{\{1,2,3,4,5\}}\) are named important1 through important5 in the Task, with additional numbered unimportant features without effect on \(y\).
library(xplainfi)
library(mlr3learners)
#> Loading required package: mlr3
task = tgen("friedman1")$generate(1000)
learner = lrn("regr.ranger", num.trees = 100)
measure = msr("regr.mse")
pfi = PFI$new(
task = task,
learner = learner,
measure = measure,
resampling = rsmp("cv", folds = 3),
n_repeats = 30
)Compute and print PFI scores:
pfi$compute()
#> ℹ Using `resample()`
pfi$importance()
#> Key: <feature>
#> feature importance
#> <char> <num>
#> 1: important1 8.183995584
#> 2: important2 7.481268675
#> 3: important3 1.571760349
#> 4: important4 12.585739572
#> 5: important5 2.810875567
#> 6: unimportant1 0.030667439
#> 7: unimportant2 -0.002837696
#> 8: unimportant3 -0.044922079
#> 9: unimportant4 -0.060054450
#> 10: unimportant5 0.060148388If it aids interpretation, importances can also be calculated as the ratio rather than the difference between the baseline and post-permutation losses:
pfi$importance(relation = "ratio")
#> Key: <feature>
#> feature importance
#> <char> <num>
#> 1: important1 2.6987668
#> 2: important2 2.5598945
#> 3: important3 1.3294180
#> 4: important4 3.6278508
#> 5: important5 1.5874860
#> 6: unimportant1 1.0067957
#> 7: unimportant2 0.9994507
#> 8: unimportant3 0.9905990
#> 9: unimportant4 0.9874657
#> 10: unimportant5 1.0126572When PFI is computed based on resampling with multiple iterations, and / or multiple permutation iterations, the individual scores can be retrieved as a data.table:
str(pfi$scores())
#> Classes 'data.table' and 'data.frame': 900 obs. of 6 variables:
#> $ feature : chr "important1" "important1" "important1" "important1" ...
#> $ iter_rsmp : int 1 1 1 1 1 1 1 1 1 1 ...
#> $ iter_repeat : int 1 2 3 4 5 6 7 8 9 10 ...
#> $ regr.mse_baseline: num 4.56 4.56 4.56 4.56 4.56 ...
#> $ regr.mse_post : num 12.3 11.9 11.3 12.1 13.6 ...
#> $ importance : num 7.77 7.33 6.74 7.56 9.06 ...
#> - attr(*, ".internal.selfref")=<externalptr>Where iter_rsmp corresponds to the resampling iteration, i.e., 3 for 3-fold cross-validation, and iter_repeat corresponds to the permutation iteration within each resampling iteration, 5 in this case. While pfi$importance() contains the means across all iterations, pfi$scores() allows you to manually visualize or aggregate them in any way you see fit.
For example:
library(ggplot2)
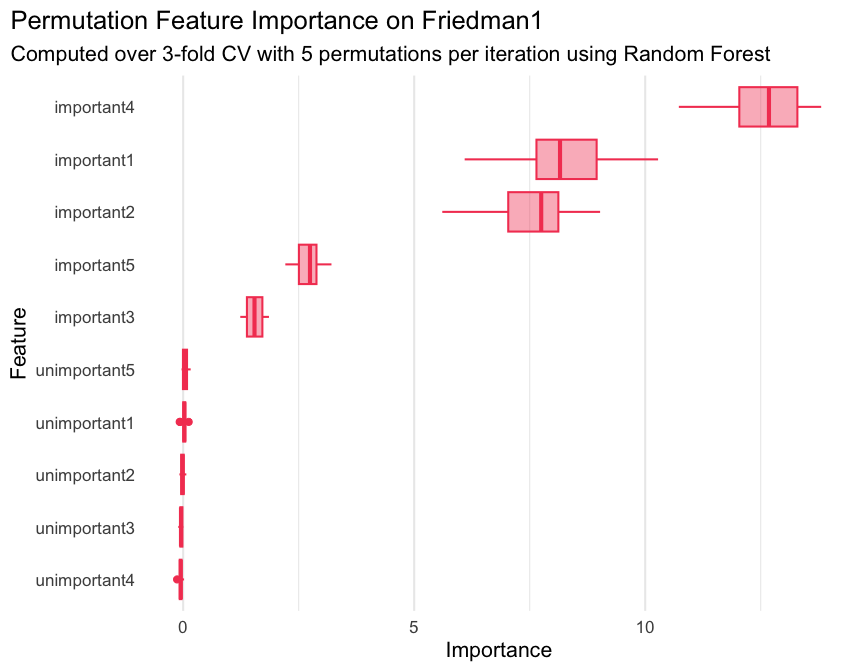
ggplot(
pfi$scores(),
aes(x = importance, y = reorder(feature, importance))
) +
geom_boxplot(color = "#f44560", fill = alpha("#f44560", 0.4)) +
labs(
title = "Permutation Feature Importance on Friedman1",
subtitle = "Computed over 3-fold CV with 5 permutations per iteration using Random Forest",
x = "Importance",
y = "Feature"
) +
theme_minimal(base_size = 16) +
theme(
plot.title.position = "plot",
panel.grid.major.y = element_blank()
)
If the measure in question needs to be maximized rather than minimized (like \(R^2\)), the internal importance calculation takes that into account via the $minimize property of the measure and calculates importances such that the intuition “performance improvement” -> “higher importance score” still holds:
pfi = PFI$new(
task = task,
learner = learner,
measure = msr("regr.rsq")
)
#> ℹ No <Resampling> provided, using `resampling = rsmp("holdout", ratio = 2/3)`
#> (test set size: 333)
pfi$compute()
#> ℹ Using `resample()`
pfi$importance()
#> Key: <feature>
#> feature importance
#> <char> <num>
#> 1: important1 0.329915393
#> 2: important2 0.297695022
#> 3: important3 0.063613087
#> 4: important4 0.493673768
#> 5: important5 0.121794662
#> 6: unimportant1 0.003972813
#> 7: unimportant2 0.002157623
#> 8: unimportant3 -0.002780577
#> 9: unimportant4 0.001914150
#> 10: unimportant5 0.001366645See vignette("xplainfi") for more examples.