library(xplainfi)
library(mlr3learners)
#> Loading required package: mlr3
library(data.table)
library(ggplot2)There are multiple (work in progress) inference methods available with the underlying implementation, but the API around them is still being worked out.
Setup
We use a simple linear DGP for demonstration purposes where
- \(X_1\) and \(X_2\) are strongly correlated (r = 0.7)
- \(X_1\) and \(X_3\) have an effect on Y
- \(X_2\) and \(X_4\) don’t have an effect
task = sim_dgp_correlated(n = 2000, r = 0.7)
learner = lrn("regr.ranger", num.trees = 500)
measure = msr("regr.mse")DAG for correlated features DGP
Variance-correction
When we calculate PFI using an appropriate resampling, such as subsampling with 15 repeats, we can use the approach recommended by Molnar et al. (2023) based on the proposed correction by Nadeau & Bengio (2003).
By default, any importance measures’ $importance()
method will not output any variances or confidence intervals, it will
merely compute averages over resampling iterations and repeats within
resamplings (iter_repeat here).
pfi = PFI$new(
task = task,
learner = learner,
resampling = rsmp("subsampling", repeats = 15),
measure = measure,
n_repeats = 20 # for stability within resampling iters
)
pfi$compute()
pfi$importance()
#> Key: <feature>
#> feature importance
#> <char> <num>
#> 1: x1 6.476555494
#> 2: x2 0.095842584
#> 3: x3 1.793989195
#> 4: x4 -0.000962437If we want unadjusted confidence intervals based on a t-distribution, we can ask for them, but note these are too narrow / optimistic and hence invalid for inference:
pfi_ci_raw = pfi$importance(ci_method = "raw", alternative = "two.sided")
pfi_ci_raw
#> Key: <feature>
#> feature importance se statistic p.value conf_lower
#> <char> <num> <num> <num> <num> <num>
#> 1: x1 6.476555494 0.0707450088 91.547879 7.519530e-21 6.32482254
#> 2: x2 0.095842584 0.0042155496 22.735490 1.879277e-12 0.08680113
#> 3: x3 1.793989195 0.0132950768 134.936355 3.311520e-23 1.76547409
#> 4: x4 -0.000962437 0.0002849623 -3.377418 4.511052e-03 -0.00157362
#> conf_upper
#> <num>
#> 1: 6.6282884472
#> 2: 0.1048840389
#> 3: 1.8225042988
#> 4: -0.0003512536The parametric CI methods ("raw" and
"nadeau_bengio") return se,
statistic, p.value, conf_lower,
and conf_upper. The alternative parameter
controls whether a one-sided test ("greater", the default,
testing H0: importance <= 0) or two-sided test
("two.sided") is performed. Here we use
alternative = "two.sided" for visualization purposes so
that conf_upper is finite.
Analogously we can retrieve the Nadeau & Bengio-adjusted standard errors and derived confidence intervals which were demonstrated to have better (but still imperfect) coverage:
pfi_ci_corrected = pfi$importance(ci_method = "nadeau_bengio", alternative = "two.sided")
pfi_ci_corrected
#> Key: <feature>
#> feature importance se statistic p.value conf_lower
#> <char> <num> <num> <num> <num> <num>
#> 1: x1 6.476555494 0.2063236235 31.390276 2.231930e-14 6.034035333
#> 2: x2 0.095842584 0.0122944003 7.795629 1.848448e-06 0.069473718
#> 3: x3 1.793989195 0.0387743032 46.267477 1.027030e-16 1.710826586
#> 4: x4 -0.000962437 0.0008310757 -1.158062 2.662136e-01 -0.002744917
#> conf_upper
#> <num>
#> 1: 6.9190756552
#> 2: 0.1222114505
#> 3: 1.8771518043
#> 4: 0.0008200431Empirical quantiles
Both "raw" and "nadeau_bengio" methods
assume normally distributed importance scores and use parametric
confidence intervals based on the t-distribution. As an alternative, we
can use empirical quantiles to construct confidence-like intervals
without parametric assumptions.
The "quantile" method returns only confidence bounds
(conf_lower, conf_upper) without
se, statistic, or p.value, as
empirical quantiles and hypothesis testing require different
methodologies.
pfi_ci_quantile = pfi$importance(ci_method = "quantile", alternative = "two.sided")
pfi_ci_quantile
#> Key: <feature>
#> feature importance conf_lower conf_upper
#> <char> <num> <num> <num>
#> 1: x1 6.476555494 6.108602488 6.8752121255
#> 2: x2 0.095842584 0.071144370 0.1213703258
#> 3: x3 1.793989195 1.736434146 1.8848348302
#> 4: x4 -0.000962437 -0.003342628 0.0006686405To highlight the differences between parametric and empirical approaches, we visualize all methods:
pfi_cis = rbindlist(
list(
pfi_ci_raw[, type := "raw"],
pfi_ci_corrected[, type := "nadeau_bengio"],
pfi_ci_quantile[, type := "quantile"]
),
fill = TRUE
)
ggplot(pfi_cis, aes(y = feature, color = type)) +
geom_errorbar(
aes(xmin = conf_lower, xmax = conf_upper),
position = position_dodge(width = 0.6),
width = .5
) +
geom_point(aes(x = importance), position = position_dodge(width = 0.6)) +
scale_color_brewer(palette = "Set2") +
labs(
title = "Parametric & non-parametric CI methods",
subtitle = "RF with 15 subsampling iterations",
color = NULL
) +
theme_minimal(base_size = 14) +
theme(legend.position = "bottom")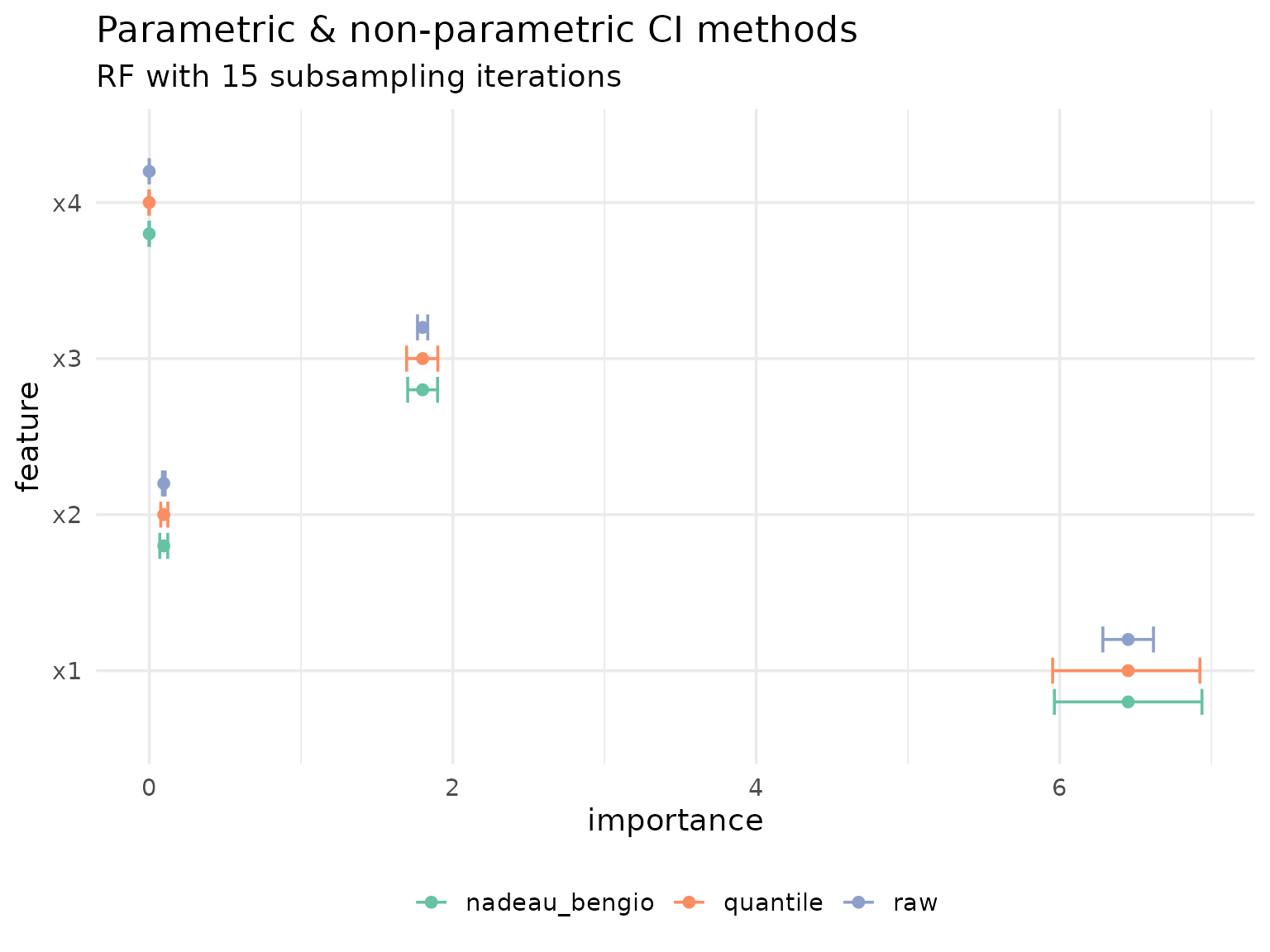
The results highlight just how optimistic the unadjusted, raw confidence intervals are.
Conditional predictive impact (CPI)
CPI (Conditional Predictive Impact) was introduced by Watson & Wright (2021) for statistical inference with conditional feature importance using knockoffs. Two main approaches are supported:
- CPI with knockoffs: The original method using model-X knockoffs for conditional sampling.
-
cARFi (Blesch et al., 2025): Uses Adversarial
Random Forests for conditional sampling, which works without Gaussian
assumptions and supports mixed data. CPI is also implemented by the cpi
package. It works with
mlr3and its output on our data looks like this:
cpi_res = cpi(
task = task,
learner = learner,
resampling = resampling,
measure = measure,
test = "t"
)
setDT(cpi_res)
setnames(cpi_res, "Variable", "feature")
cpi_res[, method := "CPI"]
cpi_res
#> feature CPI SE test statistic estimate
#> <char> <num> <num> <char> <num> <num>
#> 1: x1 4.5092929912 0.147978747 t 30.4725717 4.5092929912
#> 2: x2 0.0029567621 0.003362453 t 0.8793467 0.0029567621
#> 3: x3 1.8536055915 0.060102228 t 30.8408797 1.8536055915
#> 4: x4 -0.0008885688 0.000770307 t -1.1535255 -0.0008885688
#> p.value ci.lo method
#> <num> <num> <char>
#> 1: 3.859678e-168 4.265776760 CPI
#> 2: 1.896595e-01 -0.002576545 CPI
#> 3: 1.768282e-171 1.754700388 CPI
#> 4: 8.755837e-01 -0.002156198 CPICPI with knockoffs
Since xplainfi also includes knockoffs via the
KnockoffSampler and the
KnockoffGaussianSampler, the latter implementing the second
order Gaussian knockoffs also used by default in cpi, we
can recreate its results using CFI with the corresponding
sampler.
CFI with a knockoff sampler supports CPI inference
directly via ci_method = "cpi":
knockoff_gaussian = KnockoffGaussianSampler$new(task)
cfi = CFI$new(
task = task,
learner = learner,
resampling = resampling,
measure = measure,
sampler = knockoff_gaussian
)
#> Requested `n_repeats = 30` permutations with <KnockoffGaussianSampler>
#> ! A <KnockoffSampler> was constructed with 1 iterations
#> ℹ Proceeding with `n_repeats = 1`
#> ℹ Reconstruct <KnockoffGaussianSampler> with `iters >= 30` or use
#> <ConditionalARFSampler> if repeated sampling is required.
cfi$compute()
# CPI uses observation-wise losses; default is one-sided test (alternative = "greater")
cfi_cpi_res = cfi$importance(ci_method = "cpi")
cfi_cpi_res
#> Key: <feature>
#> feature importance se statistic p.value conf_lower
#> <char> <num> <num> <num> <num> <num>
#> 1: x1 4.362355571 0.1355320358 32.1868962 8.486605e-184 4.139321851
#> 2: x2 0.001742650 0.0029223989 0.5963079 2.755185e-01 -0.003066498
#> 3: x3 1.769344462 0.0548657780 32.2485988 2.290447e-184 1.679056446
#> 4: x4 -0.000470146 0.0009370589 -0.5017251 6.920419e-01 -0.002012185
#> conf_upper
#> <num>
#> 1: Inf
#> 2: Inf
#> 3: Inf
#> 4: Inf
# Rename columns to match cpi package output for comparison
setnames(cfi_cpi_res, c("importance", "conf_lower"), c("CPI", "ci.lo"))
cfi_cpi_res[, method := "CFI+Knockoffs"]The results should be very similar to those computed by
cpi(), so let’s compare them:
rbindlist(list(cpi_res, cfi_cpi_res), fill = TRUE) |>
ggplot(aes(y = feature, x = CPI, color = method)) +
geom_point(position = position_dodge(width = 0.3)) +
geom_errorbar(
aes(xmin = CPI, xmax = ci.lo),
position = position_dodge(width = 0.3),
width = 0.5
) +
scale_color_brewer(palette = "Dark2") +
labs(
title = "CPI and CFI with Knockoff sampler",
subtitle = "RF with 5-fold CV",
color = NULL
) +
theme_minimal(base_size = 14) +
theme(legend.position = "top")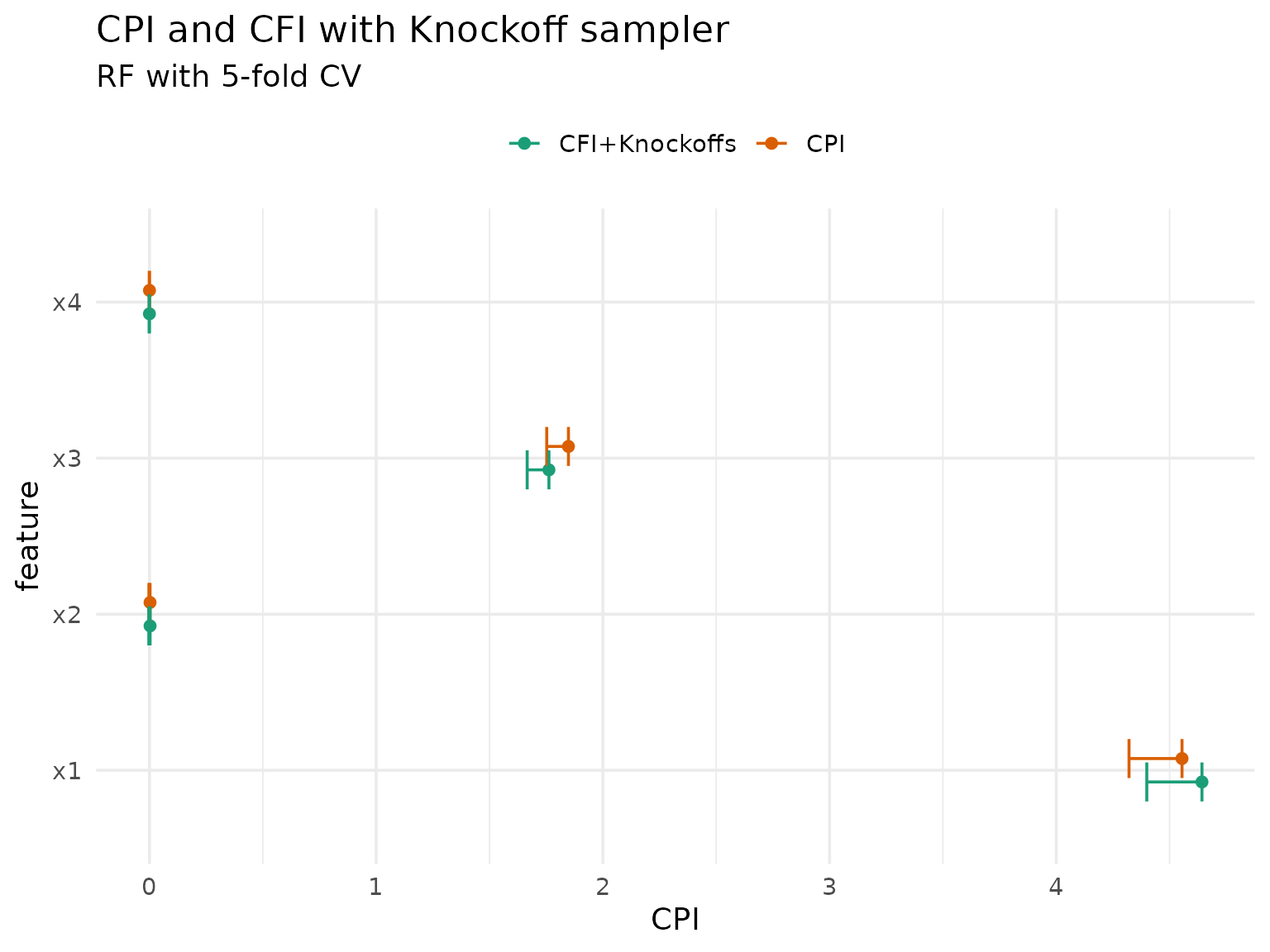
A noteable caveat of the knockoff approach is that they are not readily available for mixed data (with categorical features).
cARFi: CPI with ARF
An alternative is available using ARF as conditional sampler rather than knockoffs. This approach, called cARFi, was introduced by Blesch et al. (2025) and works without Gaussian assumptions:
arf_sampler = ConditionalARFSampler$new(
task = task,
finite_bounds = "local",
min_node_size = 20,
epsilon = 1e-15
)
cfi_arf = CFI$new(
task = task,
learner = learner,
resampling = resampling,
measure = measure,
sampler = arf_sampler
)
cfi_arf$compute()
# CPI inference with ARF sampler (cARFi)
cfi_arf_res = cfi_arf$importance(ci_method = "cpi")
cfi_arf_res
#> Key: <feature>
#> feature importance se statistic p.value conf_lower
#> <char> <num> <num> <num> <num> <num>
#> 1: x1 3.9865071718 0.0715166883 55.742335 0.00000000 3.868818148
#> 2: x2 0.0053611218 0.0025570164 2.096632 0.01807579 0.001153254
#> 3: x3 1.7527732858 0.0325190020 53.899972 0.00000000 1.699259488
#> 4: x4 -0.0009391978 0.0004918991 -1.909330 0.97181872 -0.001748675
#> conf_upper
#> <num>
#> 1: Inf
#> 2: Inf
#> 3: Inf
#> 4: Inf
# Rename columns to match cpi package output for comparison
setnames(cfi_arf_res, c("importance", "conf_lower"), c("CPI", "ci.lo"))
cfi_arf_res[, method := "CFI+ARF"]We can now compare all three methods:
rbindlist(list(cpi_res, cfi_cpi_res, cfi_arf_res), fill = TRUE) |>
ggplot(aes(y = feature, x = CPI, color = method)) +
geom_point(position = position_dodge(width = 0.3)) +
geom_errorbar(
aes(xmin = CPI, xmax = ci.lo),
position = position_dodge(width = 0.3),
width = 0.5
) +
scale_color_brewer(palette = "Dark2") +
labs(
title = "CPI and CFI with Knockoffs and ARF",
subtitle = "RF with 5-fold CV",
color = NULL
) +
theme_minimal(base_size = 14) +
theme(legend.position = "top")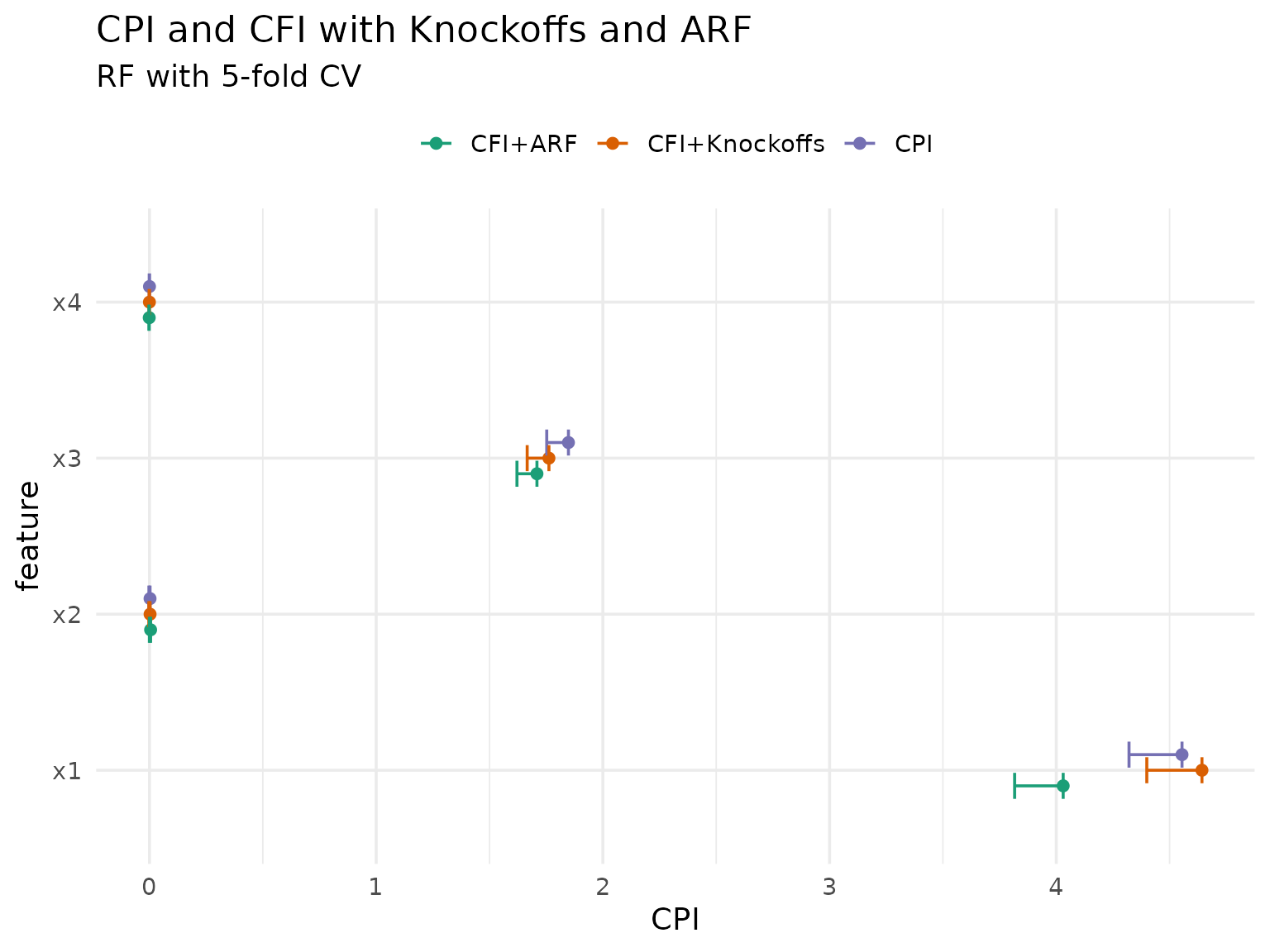
As expected, the ARF-based approach differs more from both knockoff-based approaches, but they are all roughly in agreement.
Statistical tests with CPI
CPI can also perform additional tests besides the default t-test, specifically the Wilcoxon-, Fisher-, or binomial test:
(cpi_res_wilcoxon = cfi_arf$importance(ci_method = "cpi", test = "wilcoxon"))
#> Key: <feature>
#> feature importance se statistic p.value conf_lower
#> <char> <num> <num> <num> <num> <num>
#> 1: x1 3.9865071718 0.0715166883 2001000 0.000000e+00 3.2285614819
#> 2: x2 0.0053611218 0.0025570164 1194853 2.647776e-14 0.0031150313
#> 3: x3 1.7527732858 0.0325190020 2000886 0.000000e+00 1.4036366255
#> 4: x4 -0.0009391978 0.0004918991 1031351 1.161633e-01 -0.0001004833
#> conf_upper
#> <num>
#> 1: Inf
#> 2: Inf
#> 3: Inf
#> 4: Inf
# Fisher test with same default for B as in cpi()
(cpi_res_fisher = cfi_arf$importance(ci_method = "cpi", test = "fisher", B = 1999))
#> Key: <feature>
#> feature importance se statistic p.value conf_lower
#> <char> <num> <num> <num> <num> <num>
#> 1: x1 3.9865071718 0.0715166883 3.9865071718 0.0005 3.802012761
#> 2: x2 0.0053611218 0.0025570164 0.0053611218 0.0215 0.001095074
#> 3: x3 1.7527732858 0.0325190020 1.7527732858 0.0005 1.668688790
#> 4: x4 -0.0009391978 0.0004918991 -0.0009391978 0.9670 -0.001773247
#> conf_upper
#> <num>
#> 1: Inf
#> 2: Inf
#> 3: Inf
#> 4: Inf
(cpi_res_binom = cfi_arf$importance(ci_method = "cpi", test = "binomial"))
#> Key: <feature>
#> feature importance se statistic p.value conf_lower
#> <char> <num> <num> <num> <num> <num>
#> 1: x1 3.9865071718 0.0715166883 2000 0.000000e+00 0.9985033
#> 2: x2 0.0053611218 0.0025570164 1215 2.994368e-22 0.5891816
#> 3: x3 1.7527732858 0.0325190020 1995 0.000000e+00 0.9947507
#> 4: x4 -0.0009391978 0.0004918991 1094 1.430053e-05 0.5283991
#> conf_upper
#> <num>
#> 1: 1
#> 2: 1
#> 3: 1
#> 4: 1
rbindlist(
list(
cfi_arf$importance(ci_method = "cpi")[, test := "t"],
cpi_res_wilcoxon[, test := "Wilcoxon"],
cpi_res_fisher[, test := "Fisher"],
cpi_res_binom[, test := "Binomial"]
),
fill = TRUE
) |>
ggplot(aes(y = feature, x = importance, color = test)) +
geom_point(position = position_dodge(width = 0.3)) +
geom_errorbar(
aes(xmin = importance, xmax = conf_lower),
position = position_dodge(width = 0.3),
width = 0.5
) +
scale_color_brewer(palette = "Dark2") +
labs(
title = "CPI test with CFI/ARF",
subtitle = "RF with 5-fold CV",
color = "Test"
) +
theme_minimal(base_size = 14) +
theme(legend.position = "top")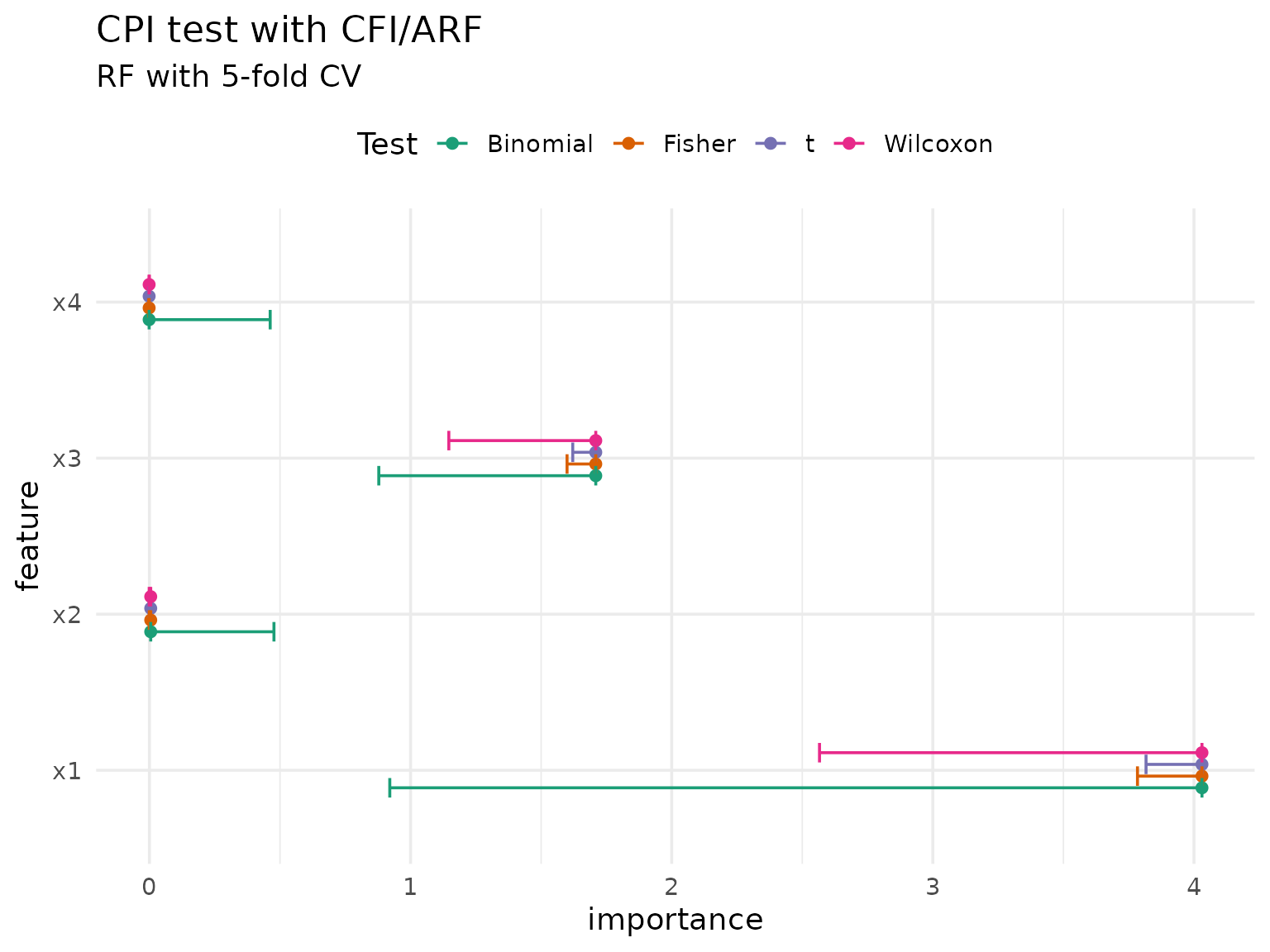
The choice of test depends on distributional assumptions: the t-test assumes normality, while Fisher and Wilcoxon are non-parametric alternatives.
Custom inference with LOCO
Lei et al. (2018) proposed inference for LOCO using the median absolute differences of the baseline- and post-refit loss differences
\[ \theta_j = \mathrm{med}\left( |Y - \hat{f}_{n_1}^{-j}(X)| - |Y - \hat{f}_{n_1}(X)| \big| D_1 \right) \]
If we apply LOCO as implemented in xplainfi
using the median absolute error (MAE) as our measure including the
median as the aggregation function, we unfortunately get something else,
though:
measure_mae = msr("regr.mae")
measure_mae$aggregator = median
loco = LOCO$new(
task = task,
learner = learner,
resampling = rsmp("holdout"),
measure = measure_mae
)
loco$compute()
loco$importance()
#> Key: <feature>
#> feature importance
#> <char> <num>
#> 1: x1 0.98413241
#> 2: x2 0.04153689
#> 3: x3 0.60559674
#> 4: x4 0.04711029This is not exactly what the authors propose,
because $score() calculates the aggregation function
(median) for each resampling iteration first, and takes the
difference afterwards, i.e.
\[ \theta_j = \mathrm{med}\left(|Y - \hat{f}_{n_1}^{-j}(X)|\right) - \mathrm{med}\left(|Y - \hat{f}_{n_1}(X)| \big| D_1 \right) \]
In the default case where the arithemtic mean is used, it does not matter whether we calculate the difference of the means or the mean of the differences, but using the median it does.
We can, however, reconstruct it by using the observation-wise losses (in this case, the absolute error):
loco_obsloss = loco$obs_loss()
head(loco_obsloss)
#> feature iter_rsmp iter_repeat row_ids loss_baseline loss_post obs_importance
#> <char> <int> <num> <int> <num> <num> <num>
#> 1: x1 1 2 1 0.06837518 0.3866730 0.31829787
#> 2: x1 1 2 3 0.33656952 0.4282341 0.09166453
#> 3: x1 1 2 14 0.14554173 1.2629784 1.11743664
#> 4: x1 1 2 18 0.21779867 0.6605524 0.44275371
#> 5: x1 1 2 25 0.07254430 1.0481222 0.97557794
#> 6: x1 1 2 28 0.22369548 1.0455213 0.82182579obs_importance here refers to the difference
loss_post - loss_baseline, so
- \(\texttt{loss_baseline} = |Y - \hat{f}_{n_1}(X)|\)
- \(\texttt{loss_post} = |Y - \hat{f}_{n_1}^{-j}(X)|\)
- \(\texttt{obs_importance} = \texttt{loss_post} - \texttt{loss_baseline}\)
Which means by taking the median for each feature \(j\) within each resampling iteration, we can construct \(\theta_j(D_1)\) as proposed, for each set \(D_k\) where \(k\) is the resampling iteration:
loco_thetas = loco_obsloss[, list(theta = median(obs_importance)), by = c("feature")]
loco_thetas
#> feature theta
#> <char> <num>
#> 1: x1 0.79280654
#> 2: x2 0.01851736
#> 3: x3 0.52224336
#> 4: x4 0.02133066The authors then propose to construct distribution-free confidence
intervals, e.g. using a sign- or Wilcoxon test We can for example use
wilcoxon.test() to compute confidence intervals around the
estimated pseudo-median:
loco_wilcox_ci = loco_obsloss[,
{
tt <- wilcox.test(
obs_importance,
conf.int = TRUE,
conf.level = 0.95
)
.(
statistic = tt$statistic,
estimate = tt$estimate, # the pseudomedian importance
p.value = tt$p.value,
conf_lower = tt$conf.int[1],
conf_upper = tt$conf.int[2]
)
},
by = feature
]
loco_wilcox_ci
#> feature statistic estimate p.value conf_lower conf_upper
#> <char> <num> <num> <num> <num> <num>
#> 1: x1 194641130 0.90926043 0.000000e+00 0.89581673 0.92270736
#> 2: x2 121302167 0.02405662 2.317954e-148 0.02219018 0.02590611
#> 3: x3 190309637 0.57687593 0.000000e+00 0.56827659 0.58544169
#> 4: x4 134365884 0.03178195 0.000000e+00 0.03022359 0.03337264